Get Started with CLASSIX
Clustering is a widely-used unsupervised learning technique to find patterns and structures in data. The applications of clustering are wide-ranging, including areas like finance, traffic, civil engineering, and bioinformatics. Clustering algorithms aim to group the data points into distinct clusters such that points within a cluster share similar characteristics on the basis of spatial properties, while points in two distinct clusters are less similar. It might be easy for a human to perceive the clusters with a small sample in a small (1 or 2) dimensional space, however, in practice, the real world data with increasing dimensions and size of the data usually make a human out of reach. Considering data provided with labels are considerably rare and expensive, reliable and easy-to-tune explainable clustering methods are highly desirable.
We introduce a novel clustering method called CLASSIX. The appealing characteristics of CLASSIX include fast speed, scalable clustering and explainable descriptions and visualization of clusters. It consists of two phases, namely a greedy aggregation phase of the sorted data into groups of nearby data points, followed by the merging of groups into clusters. The algorithm is controlled by two scalar parameters, namely a distance parameter for the aggregation and another parameter controlling the minimal cluster size. Extensive experiments are conducted to give a comprehensive evaluation of the clustering performance on synthetic and real-world datasets, with various cluster shapes and low to high feature dimensionality. Our experiments demonstrate that CLASSIX competes with state-of-the-art clustering algorithms. The algorithm has linear space complexity and achieves near linear time complexity on a wide range of problems. Its inherent simplicity allows for the generation of intuitive explanations of the computed clusters. This documentation will provide you with a beginner-friendly tutorial, and show you how to get a quick start!
Installation guide
To ensure most users can install it on their local machine, we try to keep the installing requirements for CLASSIX as low as possible. CLASSIX has the following dependencies for its clustering functionality:
cython (recommend >=0.27)
numpy>=1.7.2 (recommend >=1.20)
scipy
requests
and requires the following packages for data visualization:
matplotlib
pandas
If you want to compare the speed with other clustering algorithms in scikit-learn or other packages combined with Cython, please use CLASSIX of Cython installation for a fair comparison.
pip
To install the current release via PIP use:
pip install classixclustering
To check the installation, simply run:
python -m pip show classixclustering
If you want to uninstall it, you can use:
pip uninstall classixclustering
conda
For conda users, to install this package with conda run:
conda install -c conda-forge classixclustering
To check the installation, run:
conda list classixclustering
and uninstall it with
conda uninstall classixclustering
Installing classixclustering from the conda-forge channel can also be achieved by adding conda-forge to your channels with:
conda config --add channels conda-forge
conda config --set channel_priority strict
Once the conda-forge channel has been enabled, classixclustering can be installed with conda:
conda install classixclustering
or with mamba:
mamba install classixclustering
It is possible to list all of the versions of classixclustering available on your platform with conda:
conda search classixclustering --channel conda-forge
or with mamba:
mamba search classixclustering --channel conda-forge
Alternatively, mamba repoquery may provide more information:
# Search all versions available on your platform: mamba repoquery search classixclustering --channel conda-forge # List packages depending on classixclustering: mamba repoquery whoneeds classixclustering --channel conda-forge # List dependencies of classixclustering: mamba repoquery depends classixclustering --channel conda-forge
download
Download this repository via:
git clone https://github.com/nla-group/classix.git
If you have any installing issues, please be free to submit your questions in the issues.
Note
You can check if you Cython is installed properly:
import classix; classix.cython_is_available()
If you want to disable Cython, or compare the runtime between Cython and Python, you can simply set by
import classix; classix.__enable_cython__ = False
And then your following CLASSIX implementation will disable Cython compiling. If you can Cython back, just set classix.__enable_cython__ = True.
Quick start
CLASSIX follows a similar API design as scikit-learn library. So if you are familiar with scikit-learn, you can quickly master the CLASSIX library to do a wonderful clustering. We demonstrate a toy application of CLASSIX’s clustering on simple data.
After importing the required python libraries, we generate isotropic Gaussian blobs with 2 clusters using sklearn.datasets tool. The sample is exhibited with 2 clusters of 1000 2-dimensional data. Then, we employ CLASSIX with the simple setting:
from sklearn.datasets import make_blobs
from classix import CLASSIX
X, y = make_blobs(n_samples=1000, centers=2, n_features=2, random_state=1)
clx = CLASSIX(radius=0.5, minPts=13)
clx.fit(X)
Remember
By default, CLASSIX will use sorting=pca sorting and apply group_merging="distance", i.e., distance-based merging.
After that, to get the clustering result, we just need to load clx.labels_. Also you can return the cluster labels directly by labels = clx.fit_transform(X).
Now we plot the clustering result:
import matplotlib.pyplot as plt
plt.figure(figsize=(10,10))
plt.scatter(X[:,0], X[:,1], c=clx.labels_)
plt.show()
The result is as belows:
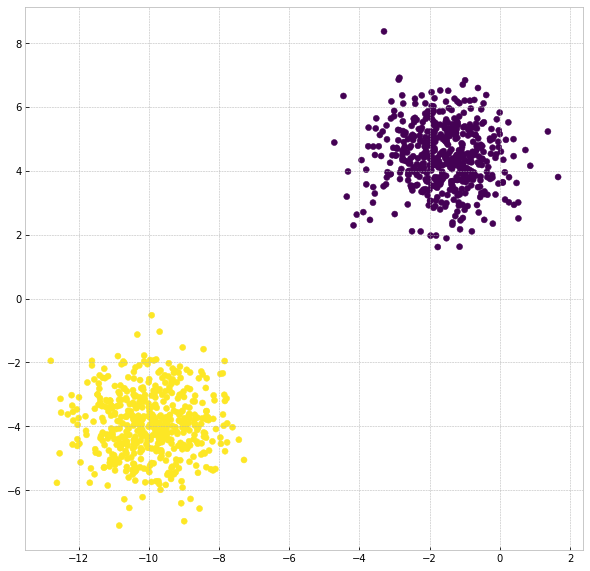
That is a basic setting tutorial of CLASSIX, which applied to most cases. If you want to learn more, please go through other sections of the documentation.
Note
CLASSIX allows to load starting points and cluster centers information by using the method clx.load_cluster_centers() and clx.load_splist_indices() where clx is the object of CLASSIX
class. More usages are shown in unittests.py